library(mlr3verse) # All the mlr3 things
library(effectplots) # For effects plotting
lgr::get_logger("mlr3")$set_threshold("error")
# Penguin Task setup
penguins <- na.omit(palmerpenguins::penguins)
penguin_task <- as_task_classif(
penguins,
target = "species"
)Feature Importance
Goals of this part:
- Introduce (some) variable importance measures
- Compare measures for different settings
1 Feature Importance
Feature importance falls under the umbrella of interpretability, which is a huge topic, and there’s a lot to explore — we’ll cover some basics and if you’re interested, you can always find more at
- The IML lecture (free slides, videos)
- The Interpretable Machine Learning book
- The
{mlr3}book chapter
Before we get started with the general methods, it should be noted that some learners bring their own method-specific importance measures. Random Forests (via {ranger}) for example has some built-in importance metrics, like the corrected Gini impurity:
lrn_ranger <- lrn("classif.ranger", importance = "impurity_corrected")
lrn_ranger$train(penguin_task)
sort(lrn_ranger$importance(), decreasing = TRUE)#> bill_length_mm flipper_length_mm bill_depth_mm island
#> 54.2401385 36.3183500 26.7463551 22.5503396
#> body_mass_g sex year
#> 21.5675075 0.8636880 -0.5303487Which shows us that for our penguins, bill_length_mm is probably the most relevant feature, whereas body_mass_g does not turn out to be as important with regard to species classification. THe year doesn’t seem to be useful for prediction at all — which is perfectly plausible!
1.1 Feature Importance with {mlr3filters}
The mlr3filters package provides some global, marginal importance methods, meaning they consider the relationship between the target and one feature at a time.
as.data.table(mlr_filters)[1:20, .(key, label)]#> Key: <key>
#> key label
#> <char> <char>
#> 1: anova ANOVA F-Test
#> 2: auc Area Under the ROC Curve Score
#> 3: boruta Burota
#> 4: carscore Correlation-Adjusted coRrelation Score
#> 5: carsurvscore Correlation-Adjusted coRrelation Survival Score
#> 6: cmim Minimal Conditional Mutual Information Maximization
#> 7: correlation Correlation
#> 8: disr Double Input Symmetrical Relevance
#> 9: find_correlation Correlation-based Score
#> 10: importance Importance Score
#> 11: information_gain Information Gain
#> 12: jmi Joint Mutual Information
#> 13: jmim Minimal Joint Mutual Information Maximization
#> 14: kruskal_test Kruskal-Wallis Test
#> 15: mim Mutual Information Maximization
#> 16: mrmr Minimum Redundancy Maximal Relevancy
#> 17: njmim Minimal Normalised Joint Mutual Information Maximization
#> 18: performance Predictive Performance
#> 19: permutation Permutation Score
#> 20: relief RELIEF
#> key labelOne “trick” of the filters package is that it can be used to access the $importance() that some learners provide on their own, and ranger provides the impurtiy-importance and permutation feature importance (PFI). We can access either with mlr3filters directly, but note it retrains the learner:
lrn_ranger <- lrn("classif.ranger", importance = "impurity_corrected")
filter_importance = flt("importance", learner = lrn_ranger)
filter_importance$calculate(penguin_task)
filter_importance#> <FilterImportance:importance>: Importance Score
#> Task Types: classif
#> Properties: missings
#> Task Properties: -
#> Packages: mlr3, mlr3learners, ranger
#> Feature types: logical, integer, numeric, character, factor, ordered
#> feature score
#> 1: bill_length_mm 50.976878008
#> 2: flipper_length_mm 36.856975328
#> 3: bill_depth_mm 25.799481552
#> 4: body_mass_g 21.751449304
#> 5: island 20.717986260
#> 6: sex 0.848159529
#> 7: year -0.003874481mlr3filters also provides a general implementation for PFI that retrains the learner repeatedly with one feature randomly shuffled.
lrn_ranger <- lrn("classif.ranger")
filter_permutation = flt("permutation", learner = lrn_ranger)
filter_permutation$calculate(penguin_task)
filter_permutation#> <FilterPermutation:permutation>: Permutation Score
#> Task Types: classif
#> Properties: missings
#> Task Properties: -
#> Packages: mlr3, mlr3learners, ranger, mlr3measures
#> Feature types: logical, integer, numeric, character, factor, ordered
#> feature score
#> 1: bill_length_mm 1.00000000
#> 2: bill_depth_mm 0.09626437
#> 3: island 0.09195402
#> 4: sex 0.08764368
#> 5: body_mass_g 0.06896552
#> 6: flipper_length_mm 0.04885057
#> 7: year 0.02442529But that also means we can use PFI for any other learner, such the SVM or XGBoost!
1.2 Your Turn!
- Compute PFI with
rangerusing it’s built-in$importance(), usingmlr3filters - Compute PFI for an SVM using the approproate
"permutation"filter - Comapre the two methods. Do they agree?
# Your codeNote that we need to use the encoding PipeOp to train the SVM of these on the penguins task, as they can’t handle categorical features automatically:
lrn_svm <- po("encode") %>>%
po("learner", lrn("classif.svm", kernel = "radial", <any other parameter>)) |>
as_learner()This is just for demonstration — we’d need to use tuned hyperparameters for the SVM for a proper comparison!
penguin_task <- as_task_classif(
na.omit(palmerpenguins::penguins),
target = "species"
)
lrn_svm <- po("encode") %>>%
po("learner", lrn("classif.svm")) |>
as_learner()
pfi_svm = flt("permutation", learner = lrn_svm)
pfi_svm$calculate(penguin_task)
pfi_svm#> <FilterPermutation:permutation>: Permutation Score
#> Task Types: classif
#> Properties: missings
#> Task Properties: -
#> Packages: mlr3, mlr3pipelines, stats, mlr3learners, e1071, mlr3measures
#> Feature types: logical, integer, numeric, character, factor, ordered,
#> POSIXct, Date
#> feature score
#> 1: bill_length_mm 1.00000000
#> 2: island -0.03846154
#> 3: bill_depth_mm -0.07122507
#> 4: sex -0.07122507
#> 5: flipper_length_mm -0.08974359
#> 6: body_mass_g -0.09116809
#> 7: year -0.10113960pfi_ranger = flt("importance", learner = lrn("classif.ranger", importance = "permutation"))
pfi_ranger$calculate(penguin_task)
pfi_ranger#> <FilterImportance:importance>: Importance Score
#> Task Types: classif
#> Properties: missings
#> Task Properties: -
#> Packages: mlr3, mlr3learners, ranger
#> Feature types: logical, integer, numeric, character, factor, ordered
#> feature score
#> 1: bill_length_mm 0.2502032220
#> 2: flipper_length_mm 0.1517506849
#> 3: bill_depth_mm 0.1254753264
#> 4: island 0.1247705530
#> 5: body_mass_g 0.0887346455
#> 6: sex 0.0130695250
#> 7: year -0.00025549211.3 Feature Effects with {effectplots}
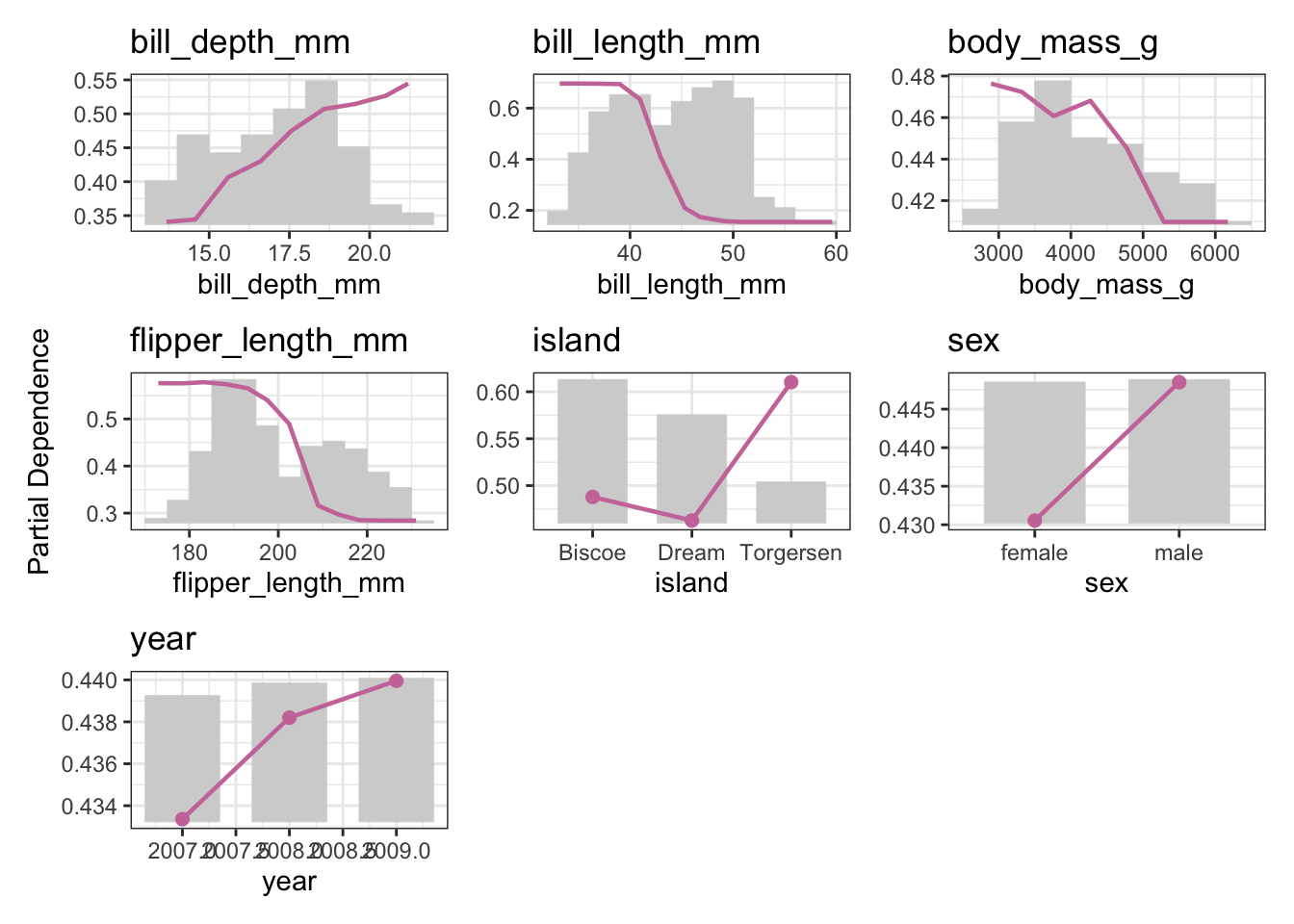
Getting a number for “is this feature important” is nice, but often we want a better picture of the feature’s effect. Think of linear models and how we can interpret \(\beta_j\) as the linear relationship between \(X_j\) and \(Y\) — often things aren’t linear though.
One approach to visualize feature effects is via Partial Dependence Plots or preferably via Accumulated Local Effect plots (ALE), which we get from the {effectplots} thankfully offers.
Let’s recycle our ranger learner and plot some effects, using the partial dependence plot (PDP) as an example:
lrn_ranger_cl <- lrn("classif.ranger", predict_type = "prob")
lrn_ranger_cl$train(penguin_task)
pd_penguins <- partial_dependence(
object = lrn_ranger_cl$model,
v = penguin_task$feature_names,
data = penguin_task$data(),
which_pred = "Adelie"
)
plot(pd_penguins)We’re doing multiclass classification here, so while our learner predicts a probability for one of each of the three target classes (Adelie, Gentoo, Chinstrap), we need to pick one for the visualization!
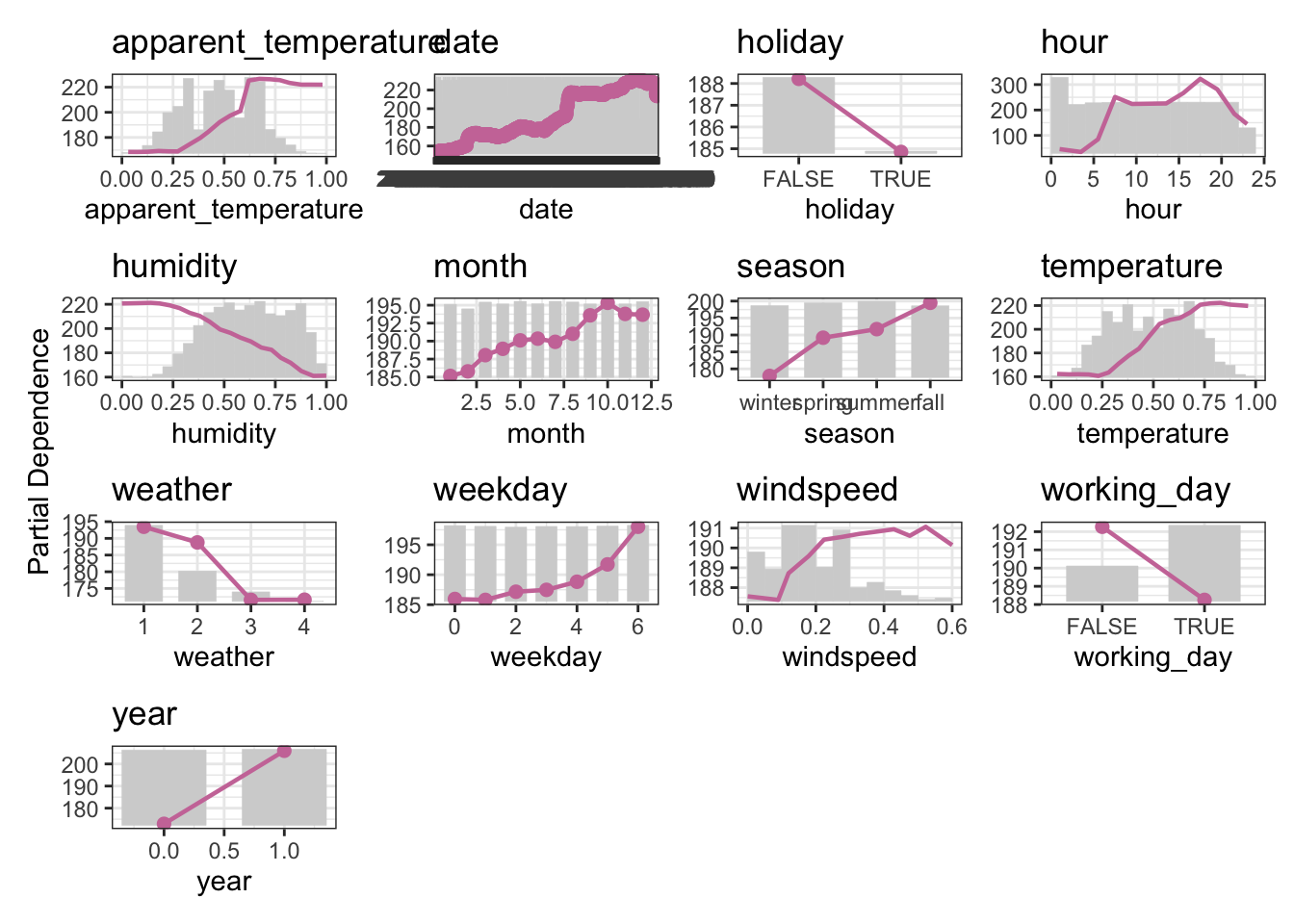
1.4 Your turn! (Possibly for another time)
- Use the
bike_shareregression task to calculate the PDP - Stick with the
rangerlearner as{effectplots}supports it directly.
# your codeThe bike_sharing task is a regression task, so make sure to switch to the regression version of the learner.
The target is bikers, the number of people using a specific bike sharing service. More information can be found on the UCI website.